Sample & Data Submission
Sample Submission For Sequencing
Our team will mail Broad barcoded tubes to your shipping contact’s address and email a corresponding merged pedigree and sample manifest spreadsheet. We understand that material can be limited, so our team can work with you if certain samples fall below recommended guidelines. All samples will be quantified upon arrival at our Center using PicoGreen or Ribogreen, and we will send those metrics back to your team prior to submitting samples for sequencing.
Each tube is pre-registered to a position in a 96-well latch rack, which we refer to as a “sample kit”. As a result of this, it is imperative that each tube remains in its pre-designated position, and that the corresponding merged pedigree and sample manifest spreadsheet be filled out accordingly. If there are extra tubes leftover after sample preparation, please discard any unused barcoded tubes, and leave any associated fields in the corresponding spreadsheet blank.
Please mail back kit(s) to the Genomics Platform at Broad at the following address:
Genomics Platform – Samples Lab
27 Blue Sky Drive
Burlington, MA 01803
Guidelines for sample quantity
Please view sample quantity guidelines for DNA, RNA, and tissue sample submissions. For other miscellaneous sample type submissions, our team will reach out with specific instructions.
DNA for exome & genome
-
3000 ng per sample
(concentrated at 50-100 ng/µl) -
Sample volume no less than
20 µL and no more than
600 µL
RNA for RNA-seq
(concentrated at 2 ng/µl)
• RQS score of at minimum of
5.5
Tissue for extraction
-
25-30 mg tissue per sample
(approximately yields
5-10 µg of RNA and 5-25 µg
of DNA )
Guidelines for Spreadsheet Entry
Sample
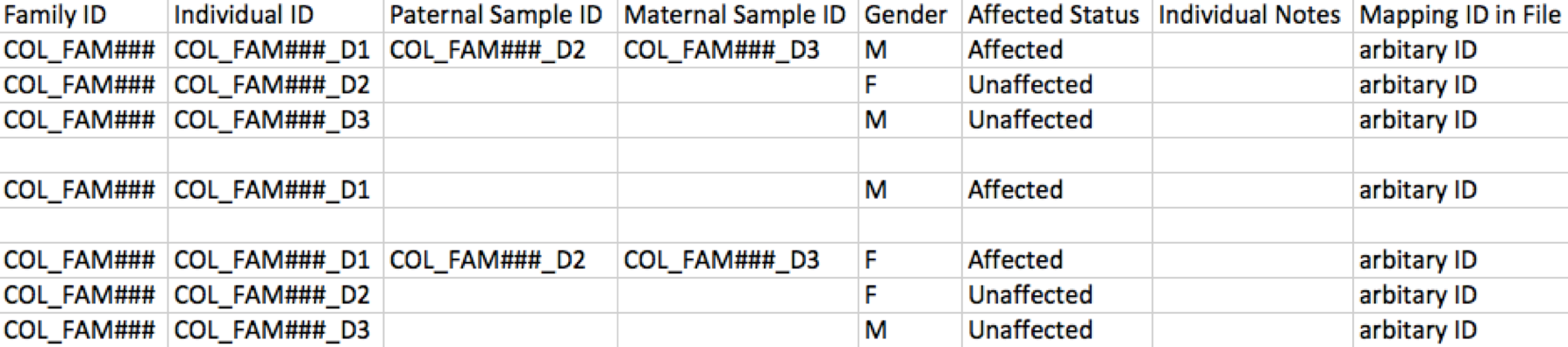
Please ensure a prefix is used as an identifier for all IDs. This prefix should be derived from the first 3 or 4 letters of the consenting PI’s last name. The collaborator sample ID should end in the aliquot number, starting with "D" for DNA samples and "R" for RNA samples. An example of pedigree entry formatting is shown below for reference.
Data
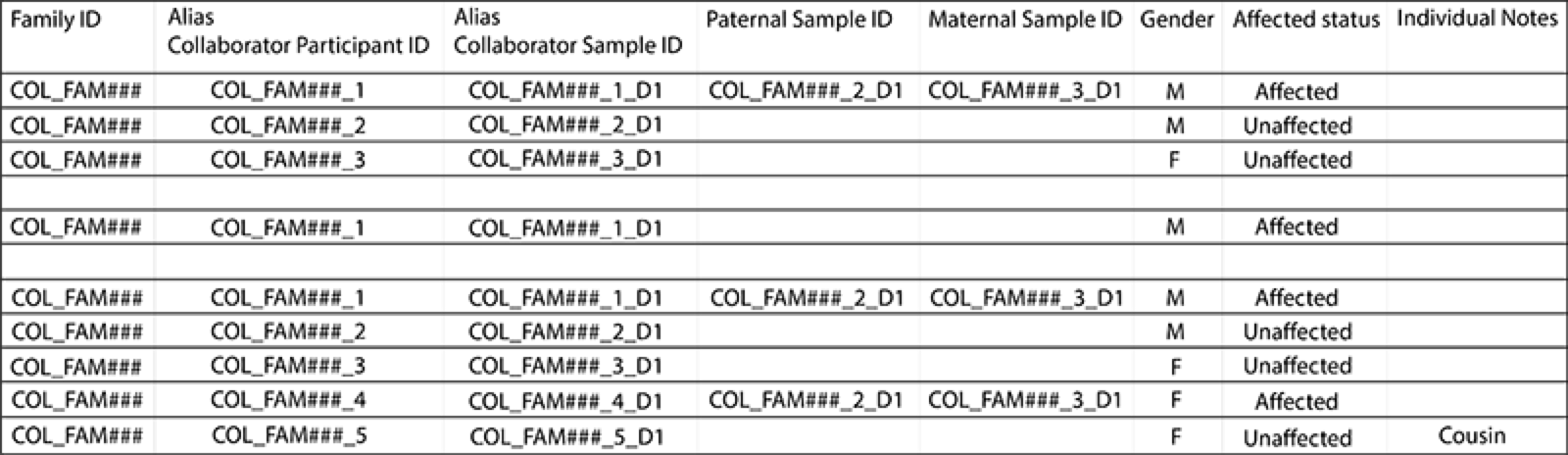
If file IDs are used as individual IDs, please ensure that an identifier is used for family IDs, which is required to be a prefix derived from the first 3 or 4 letters of the consenting PI’s last name. An example of this pedigree entry formatting is shown below for reference.
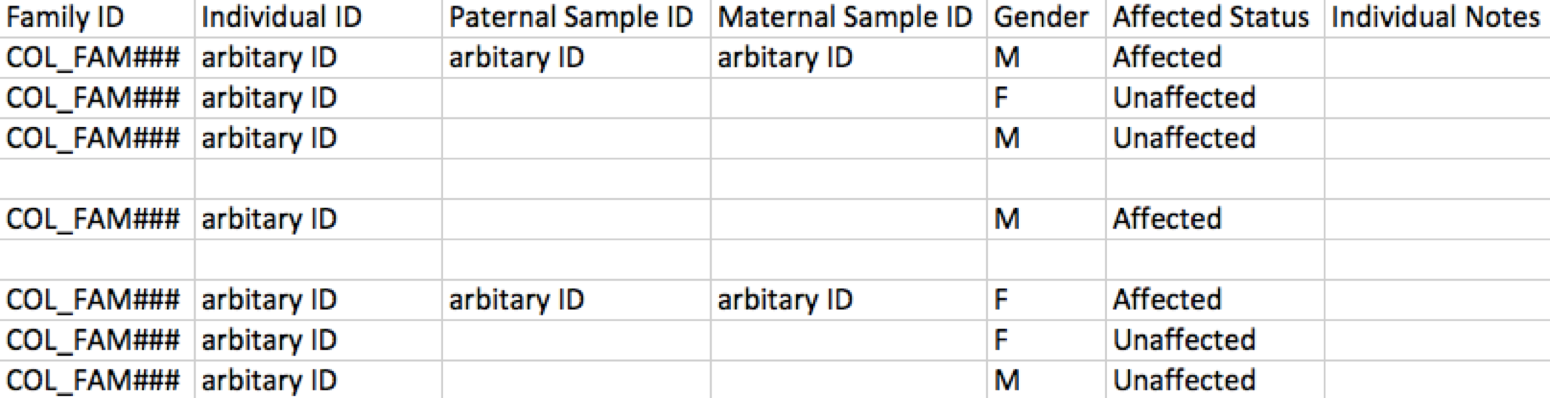
If file IDs are not used as individual IDs, please create a mapping. Additionally, ensure an identifier is used for family IDs, which is required to be a prefix derived from the first 3 or 4 letters of the consenting PI’s last name. An example of this pedigree entry formatting is shown below for reference.